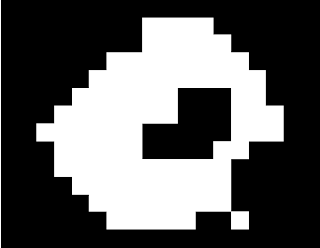
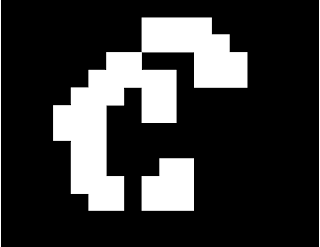The morphological operations we have learned before can be put into better use with the
opening and
closing operations. The opening operation is so called because it can "open" up spaces in an image, while the closing operation can "close" spaces depending on the structuring element (strel) used. Both operations make use of the erode and dilate operations discussed in the previous post. Let's consider this 14x18 pixel image:
To apply the opening function using a 2x2 square strel, first apply erode on the image then apply dilate on the eroded image. The resulting opened image looks like
Do you see now why the function is called
open? :)
The closing operation is just the opening operation in reverse. Dilate the image then erode the dilated image. Using the same 2x2 strel, the closed image looks closed! :D
Looking for cancer cells
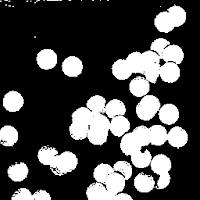
We can use the opening and closing operations to look for cancer cells (larger cells) in a pool of normal cells. Let's pretend that this image is an image of normal cells
 |
| normal cells |
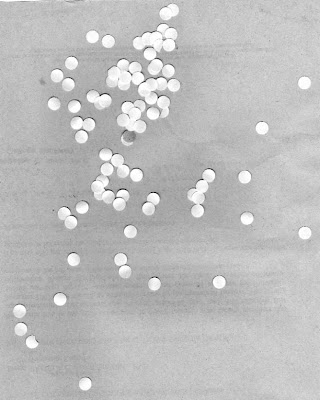
and this an image of normal cells with larger cancer cells
 |
| normal cells with cancer cells |
The goal is to separate the cancer cells from the normal cells. First we have to differentiate the normal cells from the cancer cells. We can do this by first determining the area of normal cells.
First, I cut the image into 256x256 subimages and binarized the images. Choosing carefully the threshold to separate the background from the cells. By looking at the histogram of the grayed image, we were able to determine the threshold (~0.84).
Binarized subimages.
The opening operator is then applied to separate closely placed cells. The strel used is a 5x5-pixel circle with the center at the topmost left 1-valued pixel.
Opened subimages.
Using the bwlabel function in Scilab, we can separate each white cell or blob and compute for the area of each one using follow and Green's function (see post on Area Estimation). We get the mean and standard deviation of the areas computed. The first computation returned: mean =618.53968, standard deviation=184.75568. The large standard deviation is because of the areas that are too big or too small. These are the cells that overlapped to form big white blobs or the background noise that wasn't removed with thresholding. To get the best estimate of the area of the normal cells, we must remove the outliers. I did this by removing all blobs with areas that are less than the mean+standard deviation and those that are greater than the mean+standard deviation. We can loop this until we find a mean that doesn't change much after each iteration (less than 50 pixels for me). This means that the mean we computed is the "true" mean, without the outliers. After this process, we arrive at a mean of 443.91489 pixels and standard deviation 60.940662 pixels for the normal cells.
With this, we may now look for cancer cells! Using the fact that the average area of a normal cell is ~ 444 pixels, we can make a circle with the same size and use this as strel to process the image of normal with cancer cells. First we binarize the image,
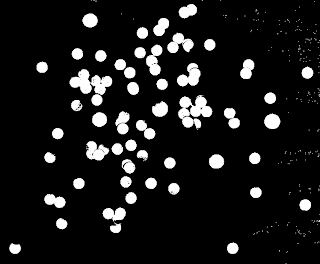 |
| Binarized image of normal cells with abnormally large cancer cells. |
Then, use the opening operation with the circular strel,
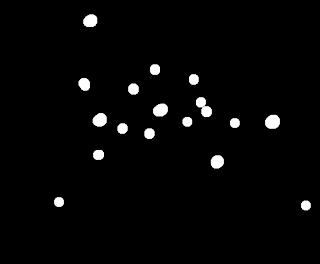 |
| Applying the opening operation on the binarized image. |
Do you see the cancer cells now? There are only 5 of them, but still too many normal cells here. Let's see if applying the opening operator again can remove these.
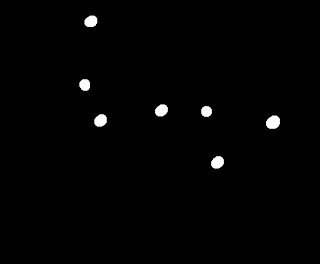 |
| The opening operation applied twice. |
Now there's the five cancer cells with two outliers. Not bad. :D
If we try to use the opening operation on this image again, we'll lose four of the cancer cells, leaving only one normal cell and one cancer cell. That's not good. Two dead normal cells are better that leaving the cancer cells behind undetected.
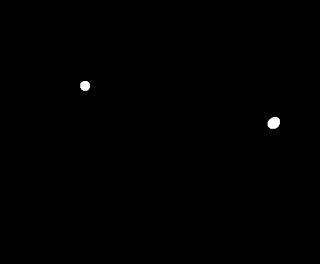 |
| The opening operator applied thrice on the binarized image. |
____________________________________________________________________________
I'm giving myself a 10 for this activity. I think I explained the opening and closing operations well enough and I liked how the rest of the blog went.